Video
2023
Artist Eduardo R. Miranda was invited to the laboratory of researcher Pablo I. Nikel at the Danish DTU Biosustain as part of the research project SinFonia. Miranda developed a method to converge the enzymes, used to handle fluorine in the cell, into musical composition. This way we can listen how the bacteria incorporates fluorine into its metabolism.
2023

Listen to the musical enzymes created by Eduardo Miranda during his residency at Pablo Nikel's lab.
1. Aldolase: https://soundclick.com/r/s8h8qm
2. Fluorinase: https://soundclick.com/r/s8h8ql
3. Aldehyde dehydrogenase: https://soundclick.com/r/s8h8qk
4. Isomerase: https://soundclick.com/r/s8h8qj
5. Kinase: https://soundclick.com/r/s8h8qi
6. Nucleosidase: https://soundclick.com/r/s8h8qh
Publications
2023

Schmidt M. (ed.) 2023. Grain & Noise: Artists in Synthetic Biology Labs – Constructive Disturbances of Art in Science. Transcript Publishing, Bielefeld
Including:
Miranda E.R. 2023. Making Music with Enzymes
Krink N, Nieto-Domínguez M & Pablo I. Rewriting the symphony of life with synthetic metabolism – Can enzymes play music?
2022
Wirth NT, Gurdo N, Krink N, Vidal-Verdú À, Donati S, Férnandez-Cabezón L, Wulff T, Nikel PI. 2022. A synthetic C2 auxotroph of Pseudomonas putida for evolutionary engineering of alternative sugar catabolic routes. Metabolic Engineering 74, https://doi.org/10.1016/j.ymben.2022.09.004
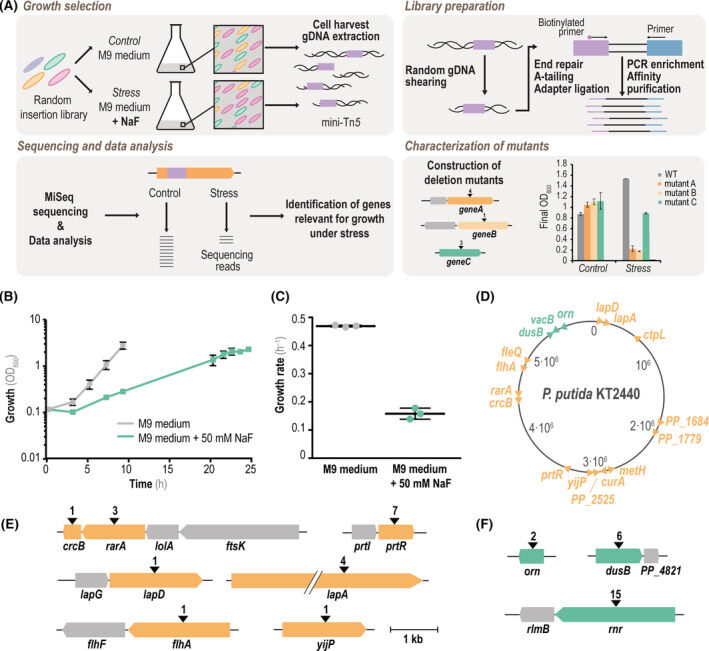
Calero P, Gurdo N, Nikel PI. 2022. Role of the CrcB transporter of Pseudomonas putida in the multi-level stress response elicited by mineral fluoride. Environmental Microbiology, https://doi.org/10.1111/1462-2920.16110
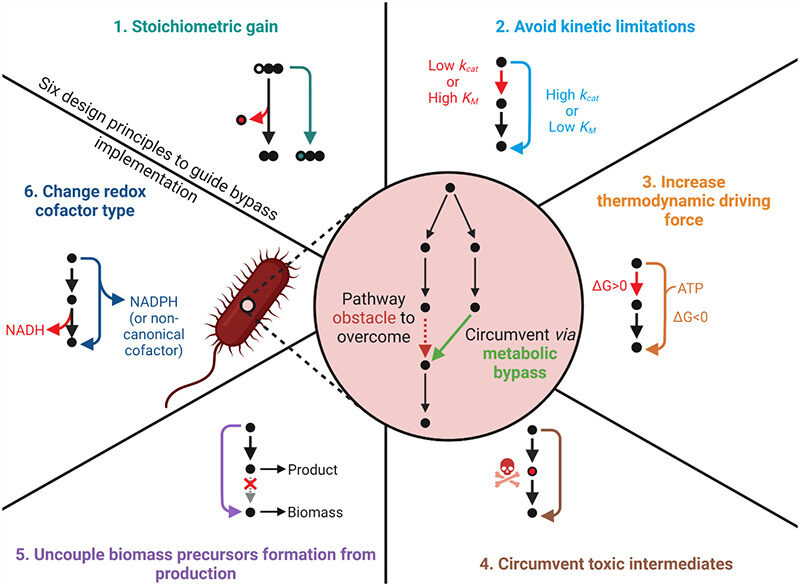
Orsi E, Claassens NJ, Nikel PI, Lindner SN. 2022. Optimizing microbial networks through metabolic bypasses. Biotechnology advances, https://doi.org/10.1016/j.biotechadv.2022.108035.
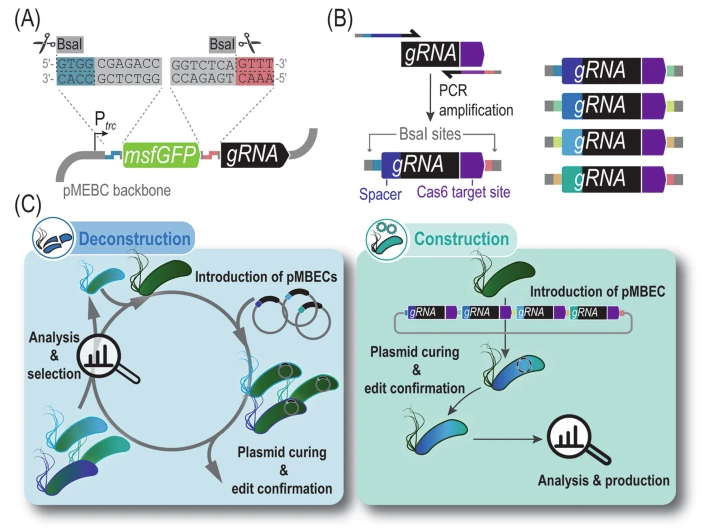
Volke DC, Martino RA, Kozaeva E, Smania AM, Nikel PI. 2022. Modular (de)construction of complex bacterial phenotypes by CRISPR/nCas9-assisted, multiplex cytidine base-editing. Nature Communications, https://doi.org/10.1038/s41467-022-30780-z
Nikel PI, Welner DH, Cros A, Volke DC. 2022. In vivo fluorine biocatalysis: Six enzymes in search of a cell factory. Chem Catalysis 2 (10), https://doi.org/10.1016/j.checat.2022.10.007

Volke DC, A Cros, DH Welner, Nikel PI. 2022. Emerging approaches for biocatalysis supporting a sustainable future: Enzymes wanted, dead or alive. Chem Catalysis 2 (10), https://doi.org/10.1016/j.checat.2022.10.006
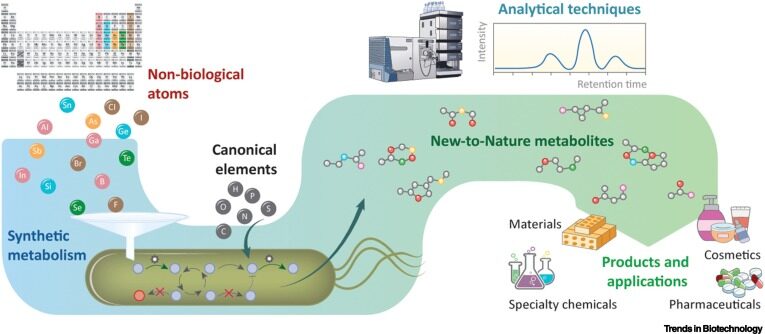
Haas R, Nikel PI. 2022. Challenges and opportunities in bringing nonbiological atoms to life with synthetic metabolism. Trends in Biotechnology. Vol 41/1: 27-45
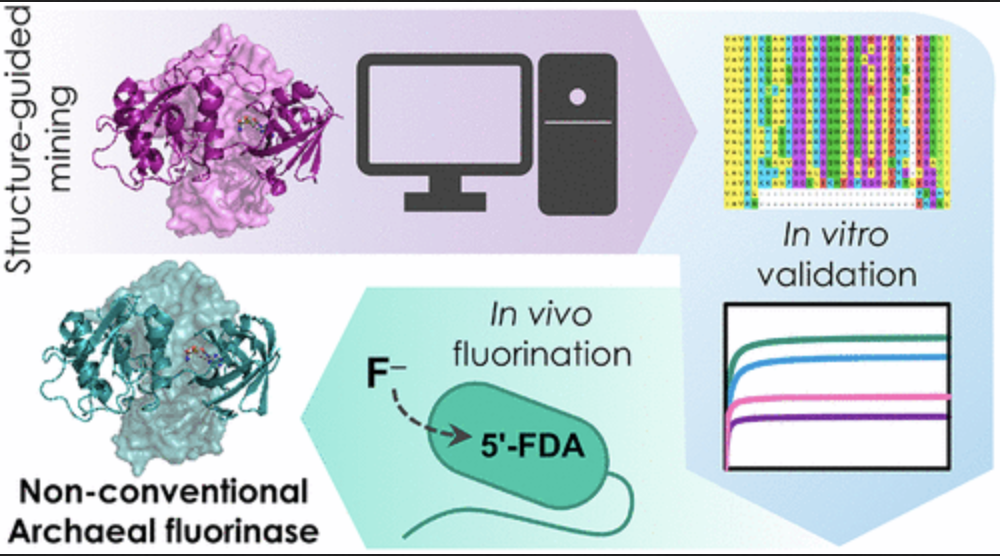
Pardo I, Bednar D, Calero P, Volke DC, Damborský J, Nikel PI. 2022.A nonconventional archaeal fluorinase identified by in silico mining for enhanced fluorine biocatalysis. ACS catalysis. Vol 12/11: 6570-6577. DOI 10.1021/acscatal.2c01184
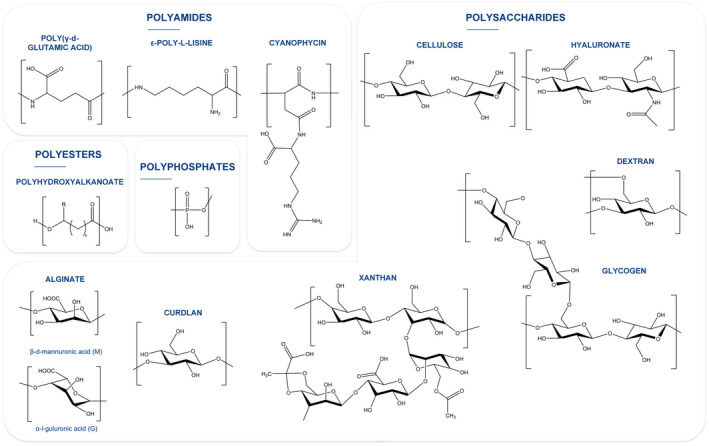
Hernández-Arriaga AM, Campano C, Rivero-Buceta V, Prieto MA. 2022. When microbial biotechnology meets material engineering. Microbial Biotechnology, https://doi.org/10.1111/1751-7915.13975
2021
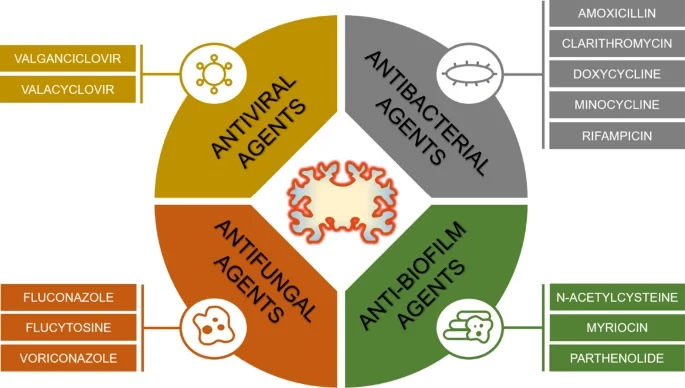
Vigasova D, Nemergut M, Liskova B, Damborsky J. 2021 Multi-pathogen infections and Alzheimer's disease. Microbial Cell Factories, https://doi.org/10.1186/s12934-021-01520-7
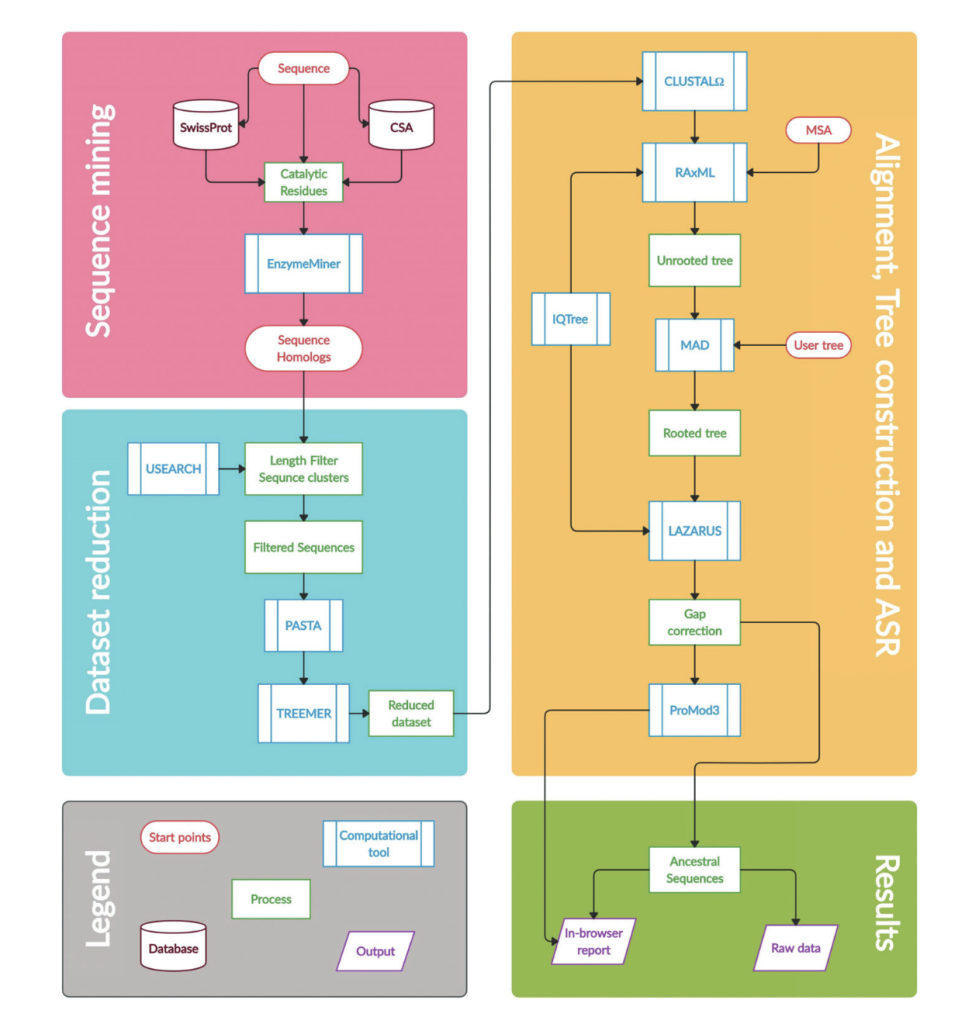
Khan RT, Musil M, Stourac J, Damborsky J, Bednar D. 2021. Fully Automated Ancestral Sequence Reconstruction using FireProtASR. Current Protocols, https://doi.org/10.1002/cpz1.30
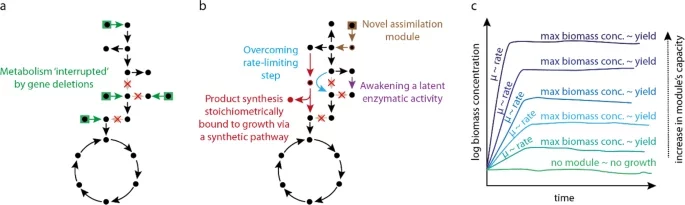
Orsi E, Claassens NJ, Nikel PI, Lindner SN. 2021. Growth-coupled selection of synthetic modules to accelerate cell factory development. Nature Communications, https://doi.org/10.1038/s41467-021-25665-6
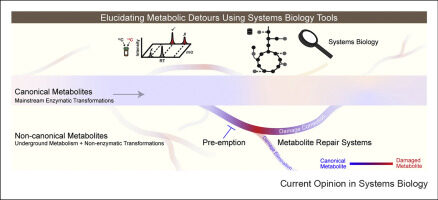
Griffith CM, Walvekar AS, Linster CL. 2021. Approaches for completing metabolic networks through metabolite damage and repair discovery. Current Opinion in Systems Biology, https://doi.org/10.1016/j.coisb.2021.100379
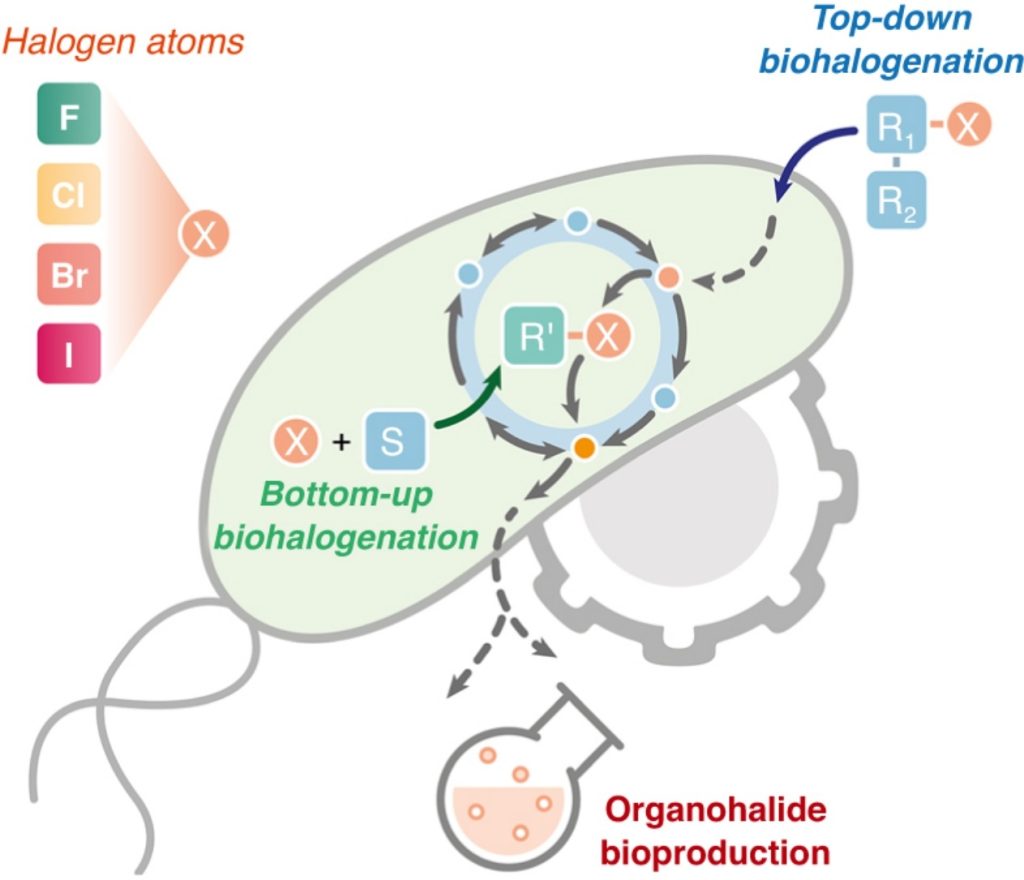
Cros A, Alfaro-Espinoza G, De Maria A, Wirth NT, Nikel PI. 2021. Synthetic metabolism for biohalogenation. Current Opinion in Biotechnology, https://doi.org/10.1016/j.copbio.2021.11.009

Marques SM, Planas-Iglesias J, Damborsky J. 2021. Web-based tools for computational enzyme design. Current Opinion in Structural Biology, https://doi.org/10.1016/j.sbi.2021.01.010
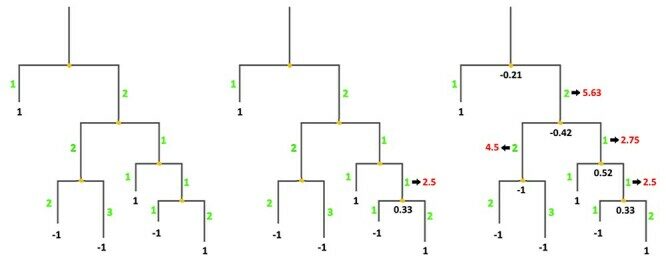
Musil M, Khan RT, Beier A, Stourac J, Konegger H, Damborsky J, Bednar D. 2021. FireProtASR: A Web Server for Fully Automated Ancestral Sequence Reconstruction. Briefings in Bioinformatics, https://doi.org/10.1093/bib/bbaa337
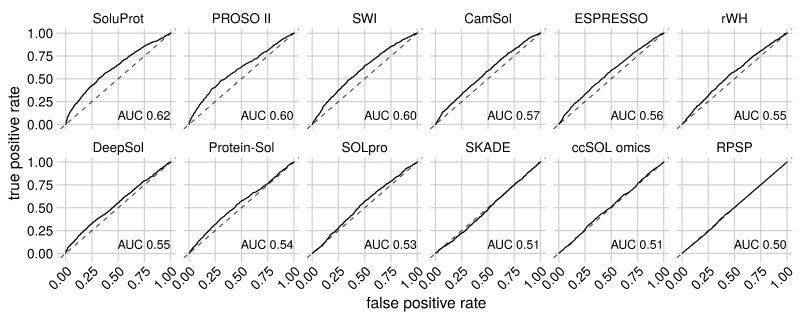
Hon J, Marusiak M, Martinek T, Kunka A, Zendulka J, Bednar D, Damborsky J. 2021. SoluProt: Prediction of Soluble Protein Expression in Escherichia coli. Bioinformatics, https://doi.org/10.1093/bioinformatics/btaa1102
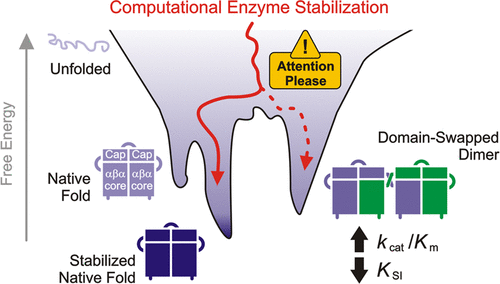
Markova K, Kunka A, Chmelova K, Havlasek M, Babkova P, Marques SM, Vasina M, Planas-Iglesias J, Chaloupkova R, Bednar D, Prokop Z, Damborsky J, and Marek M. 2021. Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers. ACS Catalysis, https://doi.org/10.1021/acscatal.1c03343
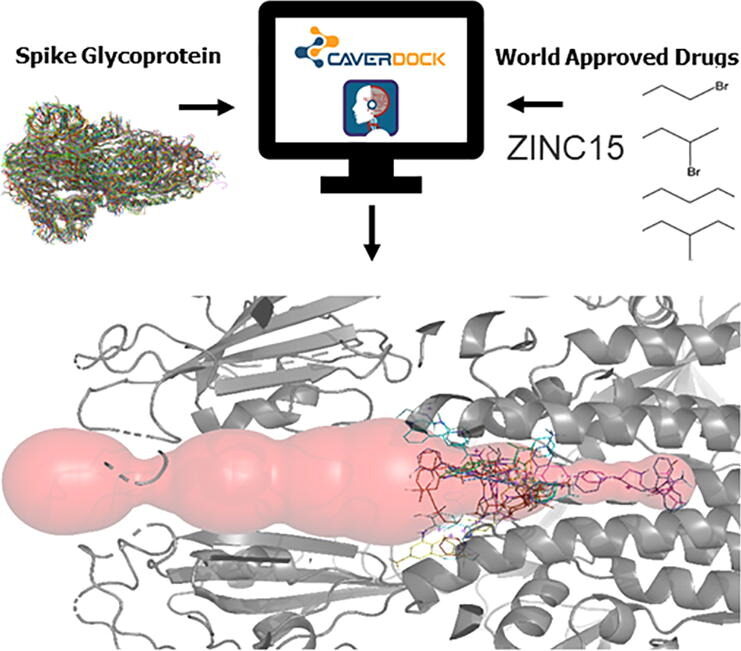
Pinto GP, Vavra O, Marques SM, Filipovic J, Bednar D, Damborsky J. 2021. Screening of world approved drugs against highly dynamical spike glycoprotein of SARS-CoV-2 using CaverDock and machine learning. Computational and Structural Biotechnology Journal, https://doi.org/10.1016/j.csbj.2021.05.043

Pérez-Pantoja D, Nikel PI, Chavarría M, de Lorenzo V. 2021. Transcriptional control of 2,4-dinitrotoluene degradation in Burkholderia sp. R34 bears a regulatory patch that eases pathway evolution. Environmental Microbiology, https://doi.org/10.1111/1462-2920.15472
2020
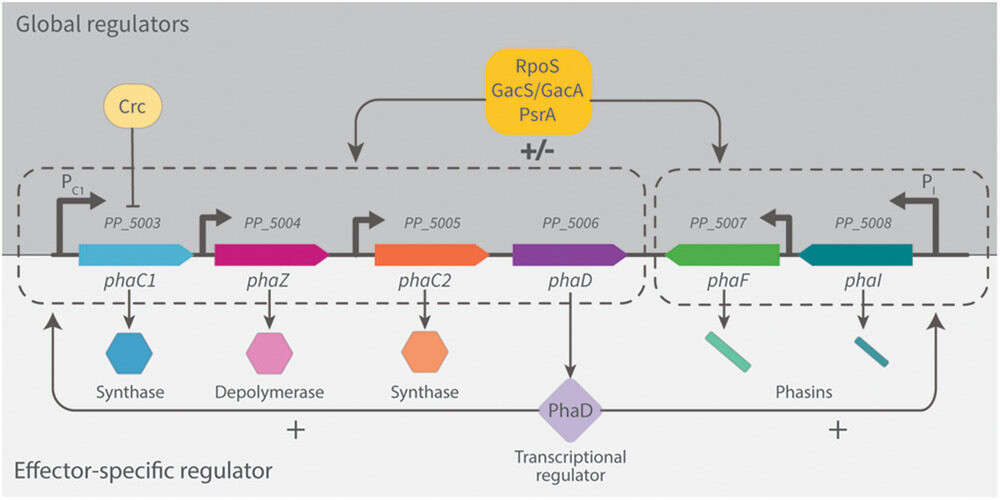
, ,, Engineering Native and Synthetic Pathways in Pseudomonas putida for the Production of Tailored Polyhydroxyalkanoates. Biotechnology Journal, https://doi.org/10.1002/biot.202000165
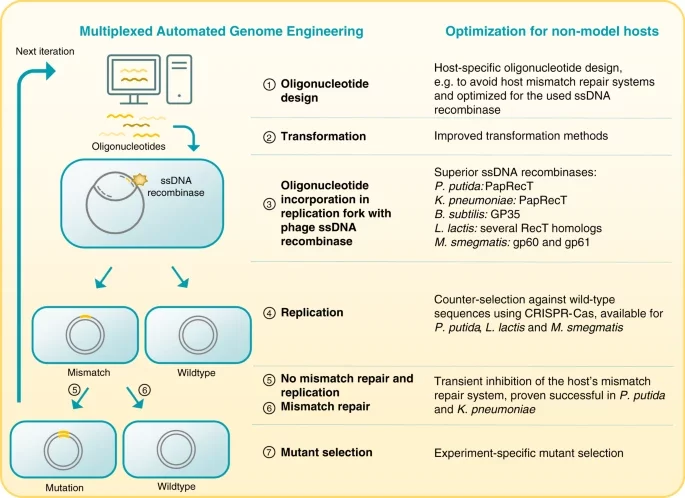
Lammens EM, Nikel PI, Lavigne R. 2020. Exploring the synthetic biology potential of bacteriophages for engineering non-model bacteria. Nature Communications, https://doi.org/10.1038/s41467-020-19124-x
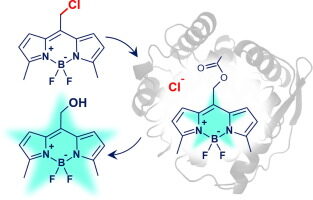
Dockalova V, Sanchez-Carnerero EM, Dunajova Z, Palao E, Slanska M, Buryska T, Damborsky J, Klán P, Prokop Z. 2020. Fluorescent substrates for haloalkane dehalogenases: Novel probes for mechanistic studies and protein labeling. Computational and Structural Biotechnology Journal, https://doi.org/10.1016/j.csbj.2020.03.029
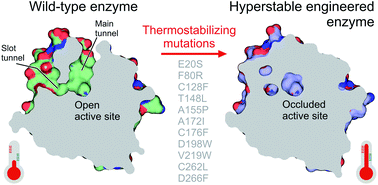
Nieto-Domínguez M, Nikel PI. Markova K, Chmelova K, Marques SM, Carpentier P, Bednar D, Damborsky J, Marek M. 2020. Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst. Chemical Science, https://doi.org/10.1039/D0SC03367G
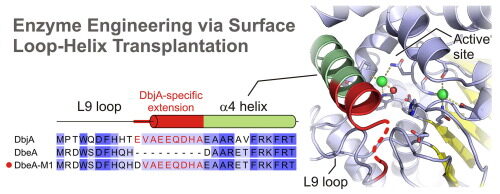
Marek M, Chaloupkova R, Prudnikova T, Sato Y, Rezacova P, Nagata Y, Kuta Smatanova I, Damborsky J. 2020. Structural and catalytic effects of surface loop-helix transplantation within haloalkane dehalogenase family. Computational and Structural Biotechnology Journal, https://doi.org/10.1016/j.csbj.2020.05.019
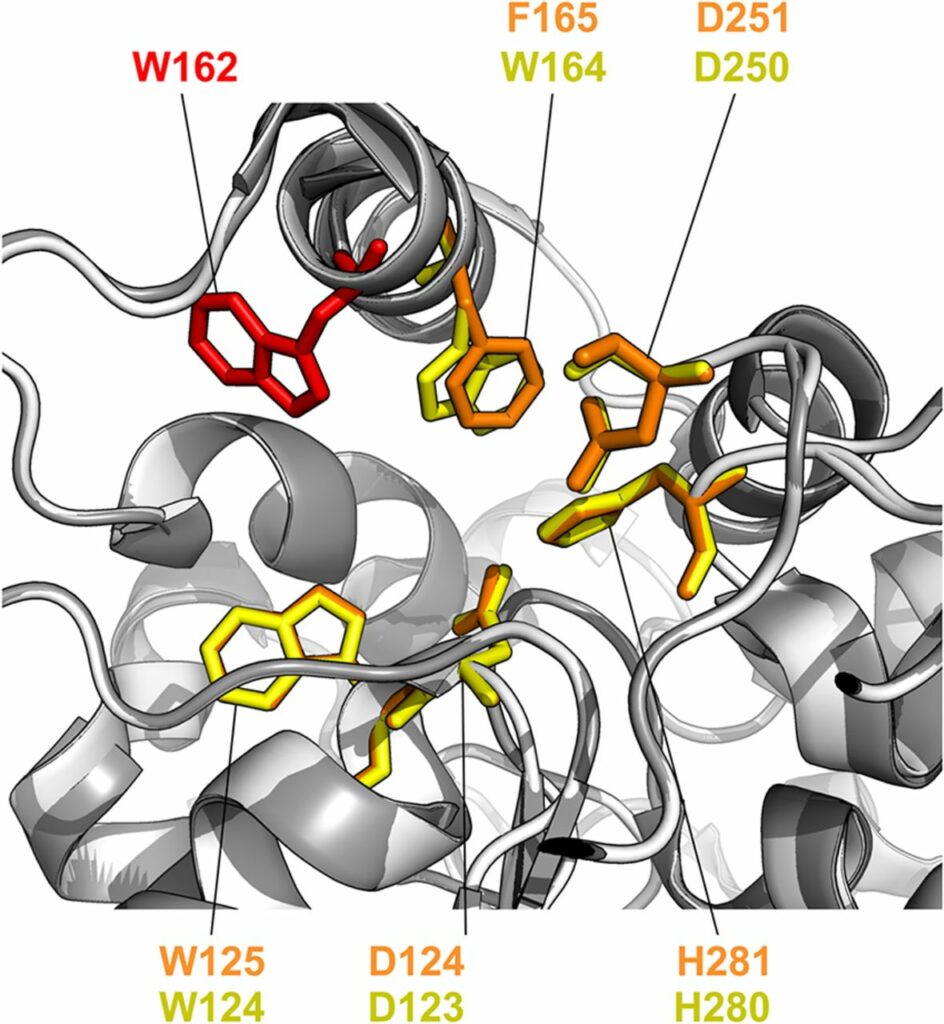
Chmelova K, Sebestova E, Liskova V, Beier A, Bednar D, Prokop Z, Chaloupkova R, Damborsky J. A 2020. Haloalkane Dehalogenase from Saccharomonospora viridis Strain DSM 43017, a Compost Bacterium with Unusual Catalytic Residues, Unique (S)-Enantiopreference, and High Thermostability. Applied and Environmental Microbiology, https://doi.org/10.1128/AEM.02820-19
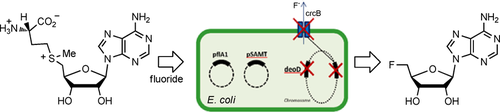
Markakis K, Lowe PT, Davison-Gates L, O'Hagan D, Rosser SJ, Elfick A. 2020. An Engineered E. coli Strain for Direct in Vivo Fluorination. ChemBioChem, https://doi.org/10.1002/cbic.202000051
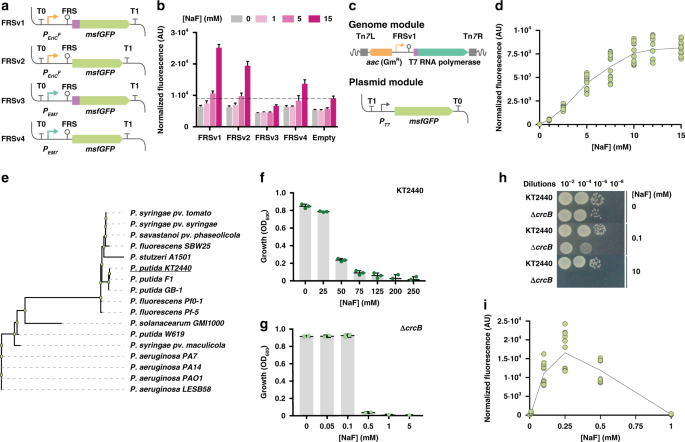
Calero P, Volke DC, Lowe PT et al. 2020. A fluoride-responsive genetic circuit enables in vivo biofluorination in engineered Pseudomonas putida. Nature Communications, https://doi.org/10.1038/s41467-020-18813-x
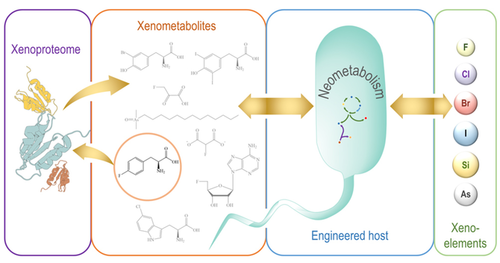
Nieto-Domínguez M, Nikel PI. 2020. Intersecting Xenobiology and Neometabolism To Bring Novel Chemistries to Life. ChemBioChem, https://doi.org/10.1002/cbic.202000091
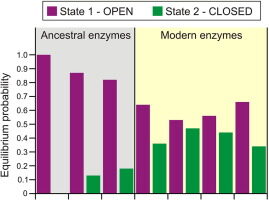
Babkova P, Dunajova Z, Chaloupkova R, Damborsky J, Bednar D, Marek M. 2020. Structures of hyperstable ancestral haloalkane dehalogenases show restricted conformational dynamics. Computational and Structural Biotechnology Journal, https://doi.org/10.1016/j.csbj.2020.06.021
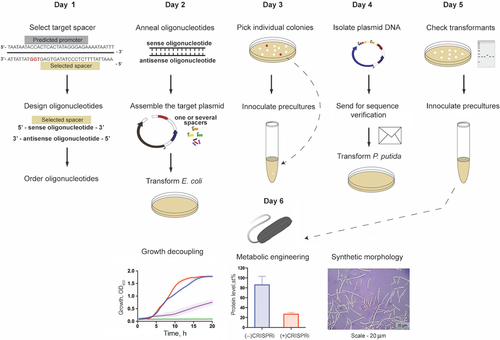
Batianis C, Kozaeva E, Damalas SG, Martín‐Pascual M, Volke DC, Nikel PI, Martins dos Santos VAP. 2020. An expanded CRISPRi toolbox for tunable control of gene expression in Pseudomonas putida. Microbial Biotechnology, https://doi.org/10.1111/1751-7915.13533

Schmidt M, Budisa N. 2020. Alternative Biofacts – Life as we don’t (yet) know it.
Chapter in the book "Art as We Don’t Know It". PDF
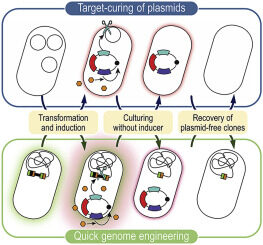
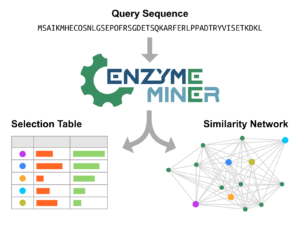
Hon J, Borko S, Stourac J, Prokop Z, Zendulka J, Bednar D, Martinek T, Damborský J. 2020.
EnzymeMiner: automated mining of soluble enzymes with diverse structures,
catalytic properties and stabilities. Nucleic Acids Research, gkaa372,
https://doi.org/10.1093/nar/gkaa372
2019
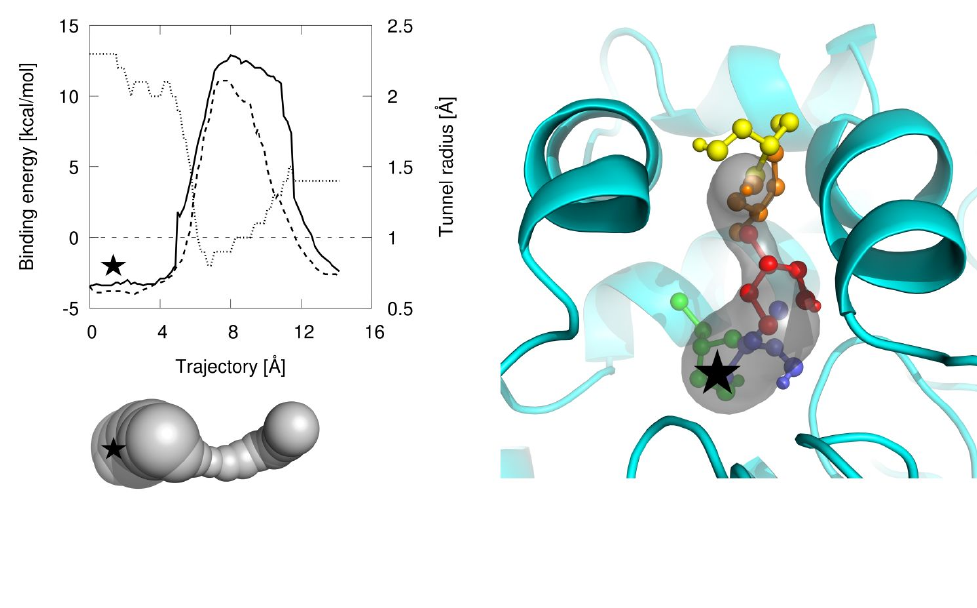
Vavra O, Filipovic J, Plhak J, Bednar D, Marques SM, Brezovsky J, Stourac J, Matyska L, Damborsky J. 2019. CaverDock: a molecular docking-based tool to analyse ligand transport through protein tunnels and channels. Bioinformatics, https://doi.org/10.1093/bioinformatics/btz386
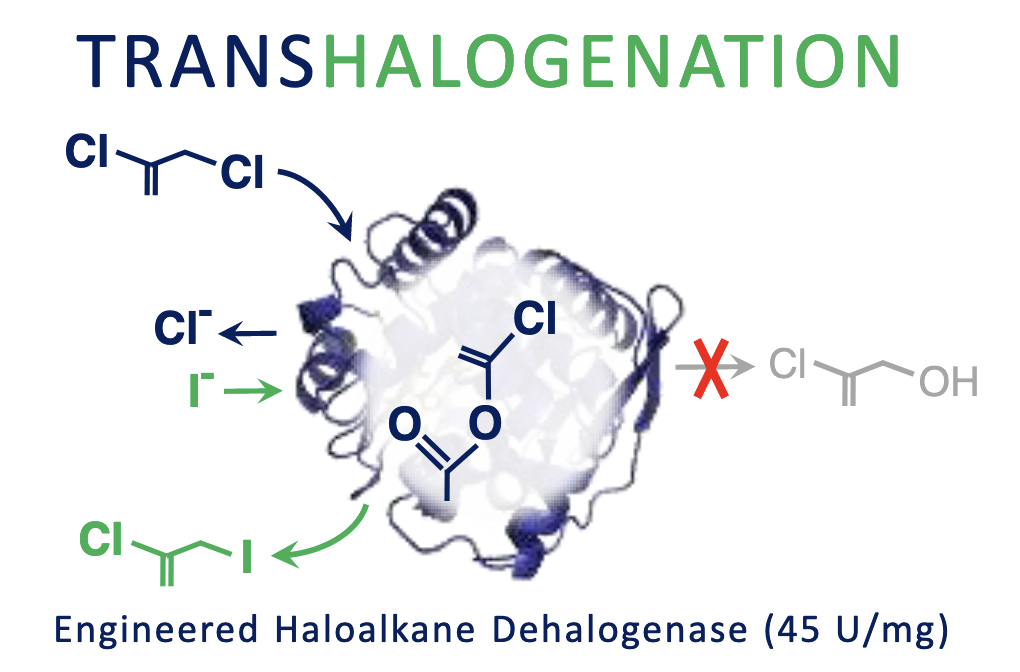
Beier A, Damborsky J, Prokop Z. 2019. Transhalogenation Catalysed by Haloalkane Dehalogenases Engineered to Stop Natural Pathway at Intermediate. Advanced Synthesis and Catalysis, https://doi.org/10.1128/AEM.02820-19
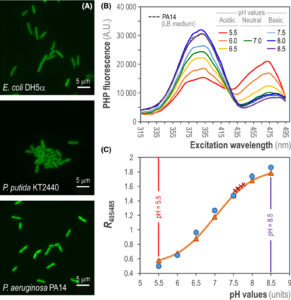
Arce‐Rodríguez A, Volke DC, Bense S, Häussler S, Nike PI. 2019. Non‐invasive, ratiometric determination of intracellular pH in Pseudomonas species using a novel genetically encoded indicator. Microbial Biotechnology Vol.12(4): 799-813. https://doi.org/10.1111/1751-7915.13439
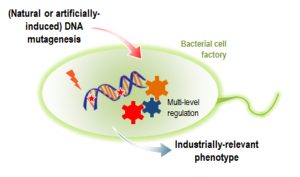
Fernández‐Cabezón L., Cros A., and Nikel PI. 2019. Evolutionary Approaches for Engineering Industrially Relevant Phenotypes in Bacterial Cell Factories. Biotechnology Journal. https://doi.org/10.1002/biot.201800439
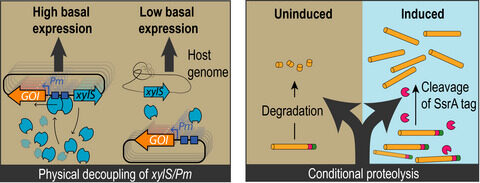
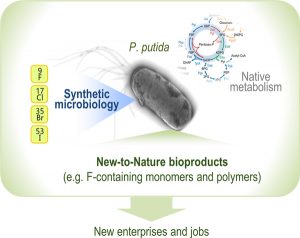
Martinelli L, Nikel PI. 2019. Breaking the state‐of‐the‐art in the chemical industry with new‐to‐Nature products via synthetic microbiology. Microbial Biotechnology. Vol. 12(2): 187-190. https://doi.org/10.1111/1751-7915.13372
